上海研匠生物科技有限公司
4 年
手机商铺
商家活跃:
产品热度:
- NaN
- 0.8999999999999999
- 0.8999999999999999
- 2.9
- 2.9
上海研匠生物科技有限公司
入驻年限:4 年
- 联系人:
陈经理
- 所在地区:
上海 闵行区
- 业务范围:
试剂、抗体、ELISA 试剂盒、技术服务、论文服务、细胞库 / 细胞培养
- 经营模式:
经销商 代理商 生产厂商 科研机构
技术资料/正文
外泌体蛋白质组学材料和方法(英文版)
236 人阅读发布时间:2023-09-21 17:56
Context
1. Materials and methods
1.1 Protein extraction and digestion
1.2 LC-MS/MS analysis
1.3 Identification and quantitation of proteins
1.4 Bioinformatic analysis
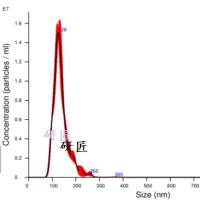
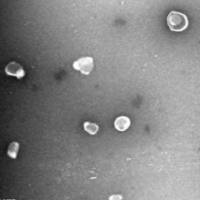
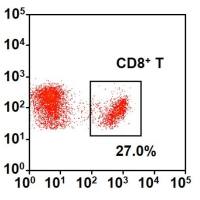
Filter-aided sample preparation (FASP Digestion) procedure:The detergent DTT (with the final concentration of 10 mM) was added to each sample respectively and mixed at 600 rpm for 1.5 h (37℃).After the samples cooled to room temperature, IAA was added with the final concentration of 20 mM into the mixture to block reduced cysteine residues and the samples were incubated for 30 min in darkness. Next, the samples were transferred to the filters respectively. The filters were washed with 100 μl UA buffer three times and then 100 μl 25mM NH4HCO3 buffer twice. Finally, trypsin was added to the samples(the trypsin : protein (wt/wt) ratio was 1:50) and incubated at 37℃ for 15-18 h (overnight), and the resulting peptides were collected as a filtrate. The peptides of each sample were desalted on C18 Cartridges (Empore™ SPE Cartridges C18 (standard density), bed I.D. 7 mm, volume 3 ml, Sigma), concentrated by vacuum centrifugation and reconstituted in 40 µl of 0.1% (v/v) formic acid. The peptide content was estimated by UV light spectral density at 280 nm using an extinctions coefficient of 1.1 of 0.1% (g/l) solution that was calculated on the basis of the frequency of tryptophan and tyrosine in vertebrate proteins.
Table Maxquant identification and quantitation indexes
Notes: Intensity-based absolute quantification (iBAQ ) and LFQ are two different methods for protein quantification provided by Maxquant software.
iBAQ Intensity reveals the level of protein expression in the sample X based on iBAQ algorithm,which is approximation to the absolute concentration of the protein in the sample.
LFQ Intensity reveals the level of protein expression in the sample X based on LFQ algorithm,which is often used in the comparison between groups.
1. Materials and methods
1.1 Protein extraction and digestion
1.2 LC-MS/MS analysis
1.3 Identification and quantitation of proteins
1.4 Bioinformatic analysis
- Materials and methods
- Protein extraction and digestion
Filter-aided sample preparation (FASP Digestion) procedure:The detergent DTT (with the final concentration of 10 mM) was added to each sample respectively and mixed at 600 rpm for 1.5 h (37℃).After the samples cooled to room temperature, IAA was added with the final concentration of 20 mM into the mixture to block reduced cysteine residues and the samples were incubated for 30 min in darkness. Next, the samples were transferred to the filters respectively. The filters were washed with 100 μl UA buffer three times and then 100 μl 25mM NH4HCO3 buffer twice. Finally, trypsin was added to the samples(the trypsin : protein (wt/wt) ratio was 1:50) and incubated at 37℃ for 15-18 h (overnight), and the resulting peptides were collected as a filtrate. The peptides of each sample were desalted on C18 Cartridges (Empore™ SPE Cartridges C18 (standard density), bed I.D. 7 mm, volume 3 ml, Sigma), concentrated by vacuum centrifugation and reconstituted in 40 µl of 0.1% (v/v) formic acid. The peptide content was estimated by UV light spectral density at 280 nm using an extinctions coefficient of 1.1 of 0.1% (g/l) solution that was calculated on the basis of the frequency of tryptophan and tyrosine in vertebrate proteins.
-
- LC-MS/MS analysis
-
- Identification and quantitation of proteins
Table Maxquant identification and quantitation indexes
| Item | Value |
| Enzyme | Trypsin |
| Max Missed Cleavages | 2 |
| Fixed modifications | Carbamidomethyl (C), |
| Variable modifications | Oxidation (M) , |
| Main search | 6 ppm |
| First search | 20ppm |
| MS/MS Tolerance | 20ppm |
| Database | xxxx for example, uniprot_mouse_76417 represents: “uniprot”, public database (http://www.uniprot.org/); “mouse”, organism species; “76417”, the number of sequences |
| Database pattern | Reverse |
| Include contaminants | True |
| protein FDR | ≤0.01 |
| Peptide FDR | ≤0.01 |
| Peptides used for protein quantification |
Use razor and unique peptides |
| Time window (match between runs) | 2min |
| protein quantification | LFQ |
| min. ratio count | 1 |
iBAQ Intensity reveals the level of protein expression in the sample X based on iBAQ algorithm,which is approximation to the absolute concentration of the protein in the sample.
LFQ Intensity reveals the level of protein expression in the sample X based on LFQ algorithm,which is often used in the comparison between groups.
-
- Bioinformatic analysis
- Cluster analysis of phosphorylated peptides
- Bioinformatic analysis
-
-
- Motif analysis
-
-
-
- Subcellular localization
-
-
-
- Domain annotation
-
-
-
- GO annotation
-
-
-
- KEGG annotation
-
注:1) 材料与方法仅供参考,不得用于正式文章发表,文章书写需自行修改。